Genomics platform enables real-time surveillance of pathogens nationally
Written by Dr Anders Gonçalves da Silva, Dr Patiyan Andersson, Associate Professor Torsten Seemann and Tuyet Hoang
The idea that rapid sharing of pathogen genomic data could significantly impact response times to outbreaks has existed for a long time – but it was the COVID-19 pandemic that highlighted the power of this kind of data sharing and expedited its implementation.
Published in Nature Communications, researchers behind the development and implementation of AusTrakka outline how the platform has facilitated rapid data sharing, democratised access to computational and bioinformatic resources and expertise, and achieved national real-time genomic surveillance.
AusTrakka is a platform designed to enable real-time sharing, analysis and reporting of pathogen genomic data across public health labs. In Australia, it is being used for rapid sharing and analysis of SARS-CoV-2 genomic sequences. It has been utilised by public health laboratories throughout the COVID-19 response to track and trace the virus across Australia and New Zealand.
The first large-scale experiment with sharing genomic data for public health started in the early 2010s led by the Food and Drug Administration (FDA) USA, in partnership with the National Center for Biotechnology Information (NCBI). The FDA was interested in rapidly comparing pathogen genomes obtained from food samples to those obtained from human clinical samples. By identifying matches quickly, point-sources of outbreaks could be identified faster, and interventions could be carried out sooner in order to stop outbreaks before they had a significant economic, social and health impact.
Other approaches have been experimented with, but their public health impact has been met with mixed success. In 2016, the Communicable Diseases Genomics Network (CDGN) and the Public Health Laboratory Network (PHLN) developed the report ‘Data sharing to improve decision making in public health: a case for Australian Public Health Laboratories’ to the Commonwealth Department of Health outlining how a system could be devised that would deliver all the potential outcomes but without the pitfalls that hindered other efforts. One crucial element to the success of such a platform was thought to be that it must exist as part of the public health system, deeply integrated into it, and not as an ad hoc feature.
Development of the platform, called AusTrakka, led by Dr Anders Gonçalves da Silva, Senior Bioinformatician, Microbiological Diagnostic Unit Public Health Laboratory (MDU PHL), Doherty Institute and Associate Professor Torsten Seemann, Lead Bioinformatician, MDU PHL, Doherty Institute, was underway before the COVID-19 global pandemic.
It was devised by public health labs for public health labs, and is embedded in the public health system, having been endorsed by PHLN and the Australian Health Protection Principal Committee (AHPCC) in 2020, and governed by CDGN.
When AusTrakka was originally conceived, it was specified that it needed to provide a central, secure, location for public health laboratories (PHLs) to deposit whole-genome sequence data of pathogens (initially, bacterial pathogens were the focus). The purpose is to find genomic matches that are close enough to suggest an epidemiological investigation is warranted.
A fundamental element of requiring a centralised system is that it ensures harmonised analyses of the data and provides access to all PHLs to the same computational resources and environment. The analytical pipelines are validated and accredited, thus providing the same high-quality analytical output to all participating PHLs. The harmonised analyses are a cornerstone of enabling consistent interpretations and responses across jurisdictions.
When the urgent need arose for a nationally consistent and real-time approach for sharing and analysing SARS-CoV-2 genome sequences, the foundation was already available and AusTrakka development was kicked into overdrive to meet the public health needs of the pandemic response.
Today, the team of bioinformaticians and genomic epidemiologists responsible for the daily operation of AusTrakka spans across Victoria, Queensland and New South Wales, with contribution from public health laboratories in all states and territories, as well as New Zealand.
AusTrakka and the associated framework for the use of the platform has cut the red tape that plagued previous multi-state investigations, streamlining the ability to rapidly share and view national data within hours instead of days. Pathogens don’t respect borders, so unifying states and territories under a single national system expedites cooperation and collaborative investigation.
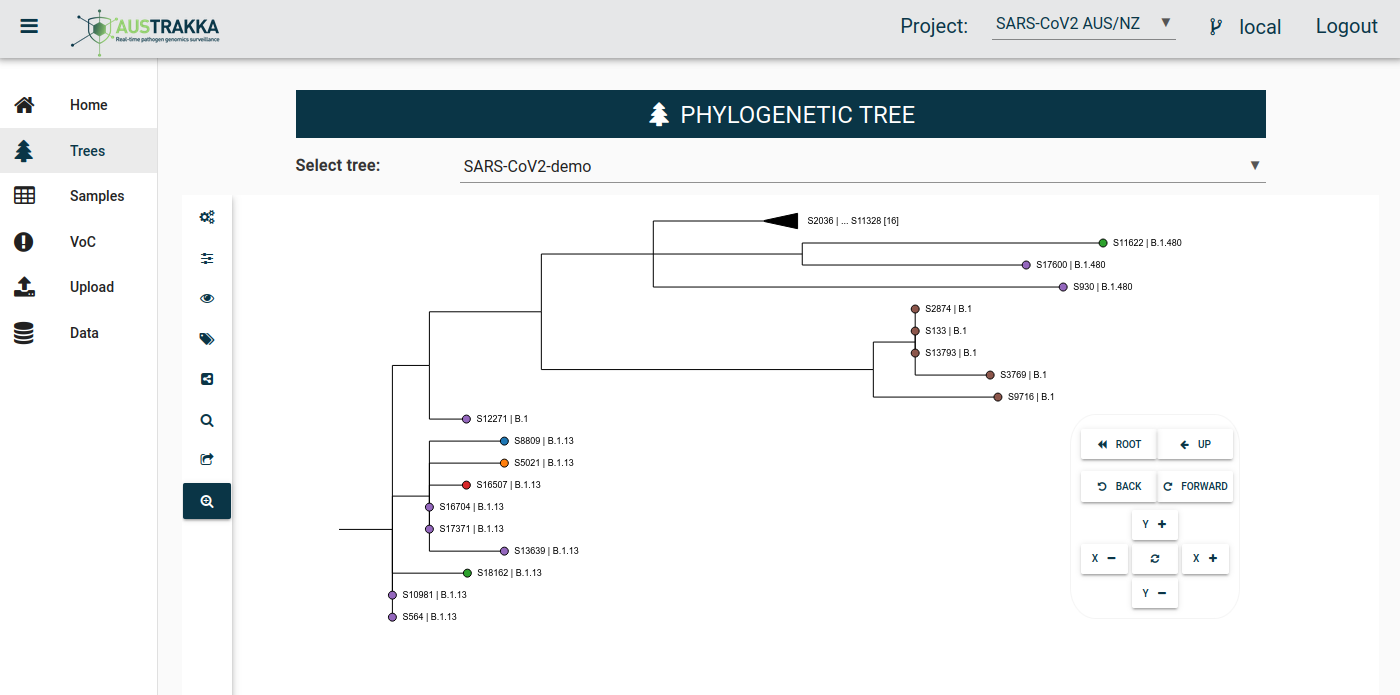
Public health laboratories across Australia upload SARS-CoV-2 genomic sequences to AusTrakka in real-time. As new data becomes available, the phylogenetic tree depicting all the sequences available for Australia and New Zealand is updated within a few hours. As soon as the new tree is available, all laboratories are able to view their sequences in the national context.
Sequences that are closely related appear as neighbouring branches on the phylogenetic tree, which allows genomic epidemiologists to quickly identify where new sequences sit relative to the historical dataset.
The data is used to track transmission within and between jurisdictions and identify new and emerging clusters and outbreaks that is used to inform public health response of COVID-19. AusTrakka also plays a critical role in the surveillance of Variants of Concern (VOC), providing frequent reporting and advice on the prevalence of VOCs to national, state and territory departments of health.
The genomic data enriches the epidemiological data by providing, for instance, strong evidence for exclusion of cases from epidemiological investigation, uncovering hidden epidemiological links, and confirming suspected epidemiological links. By enabling multi-jurisdictional data sharing and analysis, the scope of epidemiological investigations is expanded to include the whole nation.
Epidemiologists in Queensland can identify or exclude links to cases in Victoria or Western Australia, which without it could lead to extra work to track down all the cases with unknown links. By speeding up the response, it is possible to reduce the transmission of COVID-19, and better inform on the need and continuity of lockdowns and border closures.
The COVID-19 pandemic fast-tracked the establishment of a national genomics surveillance platform that has been integral to Australia’s public health surveillance and response.
As our response to the pandemic stabilises, the focus for AusTrakka will be to expand the list of pathogens covered by the platform to include additional viral and bacterial pathogens of public health importance, such as those responsible for foodborne diseases. Another front of expansion is geographic, where the aim is to include additional countries in the Southwest Pacific and Southeast Asia. By integrating international jurisdictions, we can create a regional shield to tackle outbreaks across the region and deliver better public health outcomes for Australia and the region.
The CDGN and AusTrakka team continues to work in close collaboration with the Australian Government Office of Health Protection and relevant national committees to develop nationally consistent and standardised approaches to genomic data integration, analysis and reporting of priority public health pathogens in line with the National Microbial Genomics Framework 2019-2022.
AusTrakka is funded by the Australian Department of Health and Victorian Department of Health.